巴黎多叶植物中调节多叶素生物合成的 bZIP 转录因子 PpTGA3 的鉴定
Industrial Crops and Products ( IF 5.6 ) Pub Date : 2025-01-14 , DOI: 10.1016/j.indcrop.2025.120492
Xiu-Lan Pu , Li-Yuan Zhang , Jin-Yan Zhang , Xiao Ye , Liang Lin , Dong-Rong Wu , Juan-Juan Kang , Hong Hu , Jing Chen , Kai Guo , Yue-Gui Chen , Yang Tao , Sheng-Hong Li , Yan Liu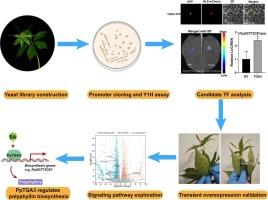
Identification of a bZIP transcription factor PpTGA3 regulating polyphyllin biosynthesis in Paris polyphylla
The biosynthesis and transcriptional regulation of active ingredients in medicinal plants have garnered significant research attention. In this study, we focused on the transcriptional regulation of polyphyllins, a group of steroidal saponins with promising pharmacological activities and industrial values in Paris polyphylla, a well-known traditional Chinese herb with significantly large genome. We constructed a yeast hybrid library for P. polyphylla var. yunnanensis (PPY) to identify transcription factors modulating polyphyllin biosynthesis. Using the promoters of two genes encoding UDP-glycosyltransferases (UGTs) involved in polyphyllin biosynthesis as baits, we isolated potential transcriptional regulators. Subsequent analyses, including subcellular localization, transcriptional activity, and dual-luciferase assays, indicated that the nuclear-localized basic region leucine zipper (bZIP) transcription factor, PpTGA3, could activate PpUGT73CE1 expression. PpTGA3, predominantly expressed in the rhizome and sepal, was found to alter polyphyllin content upon transient overexpression in P. polyphylla leaves. Furthermore, exogenous salicylic acid (SA) treatment and transcriptome analysis suggest a potential role of SA in polyphyllin accumulation, indicating an intricate interplay between SA signaling and PpTGA3. Overall, our findings provide new insights into the regulation of polyphyllin biosynthesis and establish a molecular basis for enhancing polyphyllin production in P. polyphylla.

访问手机版

微信公众号